What is cryo-EM?
Cryo-EM combines the power of electron microscopes to see tiny things and the power of cryogenic temperatures to preserve their natural shapes and structures. (Think freezing, freezing cold, around −183 °C or nearly –300 °F: more than twice as cold as the coldest surface temperature ever recorded on Earth!) A relative newcomer in the world of structural biology, cryo-EM had been nicknamed “blobology” for the indistinct, low-resolution pictures of molecular structures it produced — like the gray image below. Now, it has revolutionized the field thanks to its own “resolution revolution.”

Image: A ribosome cryo-EM model, stylized to depict the limited resolution achievable by this technique several decades ago. Video: A high-resolution ribosome model with distinct features identified by color based on Protein Data Bank entry 6QZP.
Cryo-EM can capture flexible, wiggly proteins not suited to x-ray crystallography, which has traditionally been the most common way to probe the 3D structure of biomolecules. And it can illuminate big molecules like intact viruses, giving it an advantage over NMR — short for nuclear magnetic resonance spectroscopy — a technique that can reveal information about biological structures in their natural, dynamic state but that is limited to small molecules.
Arguably the most exciting entry point to structural biology in recent years, today’s cryo-EM is known for the remarkable and ever-increasing precision with which it “solves” molecular structures, especially through its most canonical form, single-particle analysis. These feats earned cryo-EM’s early pioneers the 2017 Nobel Prize in Chemistry.
But the world of cryo-EM is also evolving in creative directions, with a variety of approaches to choose from and even combine, like cryo-electron tomography and light microscopy, allowing structural biologists to map the best possible route to scientific discovery.
Sizing up cryo-EM
How do we measure the tiny molecular machines at work inside us? And how do we measure the success of cryo-EM?
Figure by Alysa Zhou and Matt Stefely
Different kinds of microscopes and techniques (top row) illuminate biology across the scale of life (bottom row). As the overlap across the techniques in green, turquoise, blue, and purple suggests, powerful combinations of approaches are possible. The scale bar (middle row) shows the units of length we use to measure our bodies from blood vessels all the way down to atoms. While light microscopy allow us to peer into objects ranging from about a millimeter (mm), through the micrometer range (μm) and down to about 100 nanometers (nm), cryo-EM helps us see much smaller things — including viruses, proteins, and nucleic acids.
These same units of length are also used to measure resolution. Think of resolution as the smallest distance between two points that can be distinguished as separate from each other in a microscopy image. Put simply, grainy, blobby images are low resolution, while sharply defined, detailed images are high resolution.
Structural biologists often convey resolution in Ångströms (Å) — a unit that is ten billion times smaller than the meter at the far left of the scale bar, or about equivalent to the radius of a single atom. The better (or higher) the resolution, the lower the Ångström. At about 3 Å, or “near-atomic resolution,” particles in the image are resolved enough that a model of a molecule’s structure can be built by incorporating other computational techniques. Atomic resolution at or very close to 1 Å means that we can see the distance between two adjacent atoms!
Why is cryo-EM important?
In 1952, biochemist Rosalind Franklin and her PhD student Raymond Gosling used X-ray diffraction to take a now famous photograph of DNA. Known as Photograph 51, this image played a crucial role in Francis Crick and James Watson’s claim in 1953 that DNA takes the 3D shape of a double-stranded helix1. That understanding of structure then unlocked the idea that DNA functions as a crucial genetic instruction manual.
The important discoveries made possible by cryo-EM follow in this tradition of revealing “structure–function” relationships. Today scientists can work to determine not only the structure of molecules but also the changes those structures undergo during important biological processes. For example, cryo-EM has captured the way a protein called PriA opens up to set in motion a sequence of events that allows the bacteria E. coli to restart the replication of its genome even when that process gets interrupted.2

Cryo-EM’s impact also extends beyond the world of fundamental research, with huge potential to benefit the discovery and development of new drugs. Scientists at the Laboratory of Infectious Diseases at NIAID, for example, used cryo-EM to determine the 3D structure of a universal flu vaccine, known as H1ssF_3928. The NIH opened a two-year Phase 1 Clinical Trial of the vaccine in 2023.

Image: NIAID
This colorized version of the vaccine nanoparticle represents eight influenza hemagglutinin proteins (yellow) arrayed on the surface of a protein scaffold (blue).
How does cryo-EM work?
The cryo-EM workflow involves three main steps: sample preparation at the lab bench, imaging at the microscope, and image processing at the computer. At each step, structural biologists address the double-edged nature of the ionizing electron beam that illuminates — but also inevitably damages — the structure of the sample.
At the lab bench
Imagine we want to use single-particle analysis to determine the structure of a molecule — say, a ribosome, the cellular machinery that builds proteins. First, we need to purify and concentrate our sample of ribosomes, which we’ll then deposit onto a very small metal mesh grid. Extra solution surrounding the sample is blotted off before a robotic arm plunges the grid into a cryogenic liquid, usually ethane at –183 ºC. This step instantly traps the particles in the sample on the grid in a layer of a special glass-like ice called vitreous or amorphous ice.
Plunge freezing helps preserve the natural state of the molecule in part by preventing the formation of crystals, which happens when water freezes slowly and would damage the delicate molecular structures. While crystalline ice is the most common form of ice on Earth, vitreous ice is actually the most common form of ice in outer space and thus in our universe! Freezing also helps reduce the radiation damage caused by the electron beam in the next step.
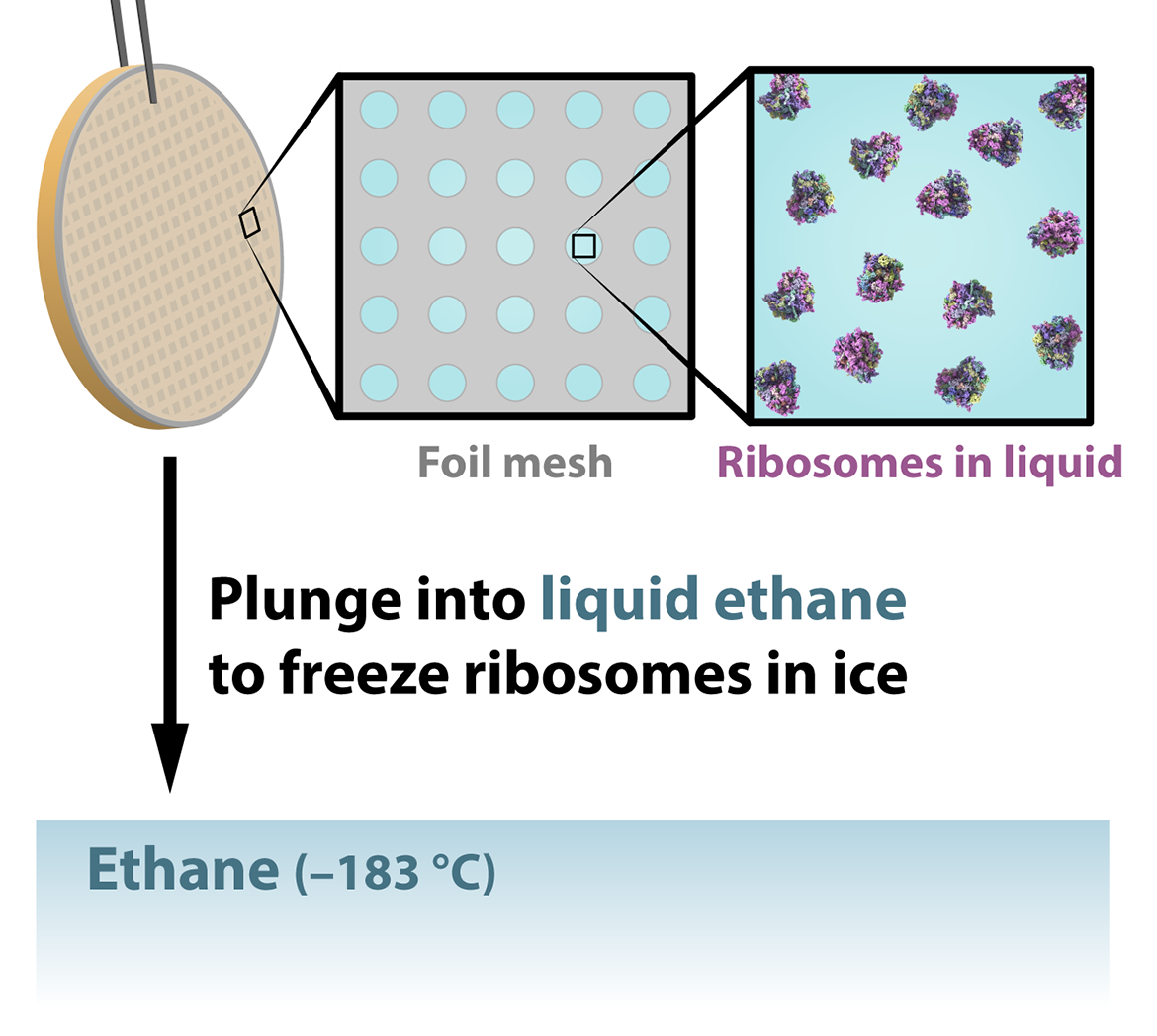
At the microscope
When the sample is flash frozen, the ribosomes trapped in the ice are ideally arranged in random orientations, so that we’ll be able to image their structures from up, down, and all around. Shot with an electron beam, these random orientations appear as 2D shadows on a detector, producing many thousands of images of different views of the particles. Contrary to its name, single-particle analysis actually requires thousands of sample particles to create views of the structure from as many angles as possible.
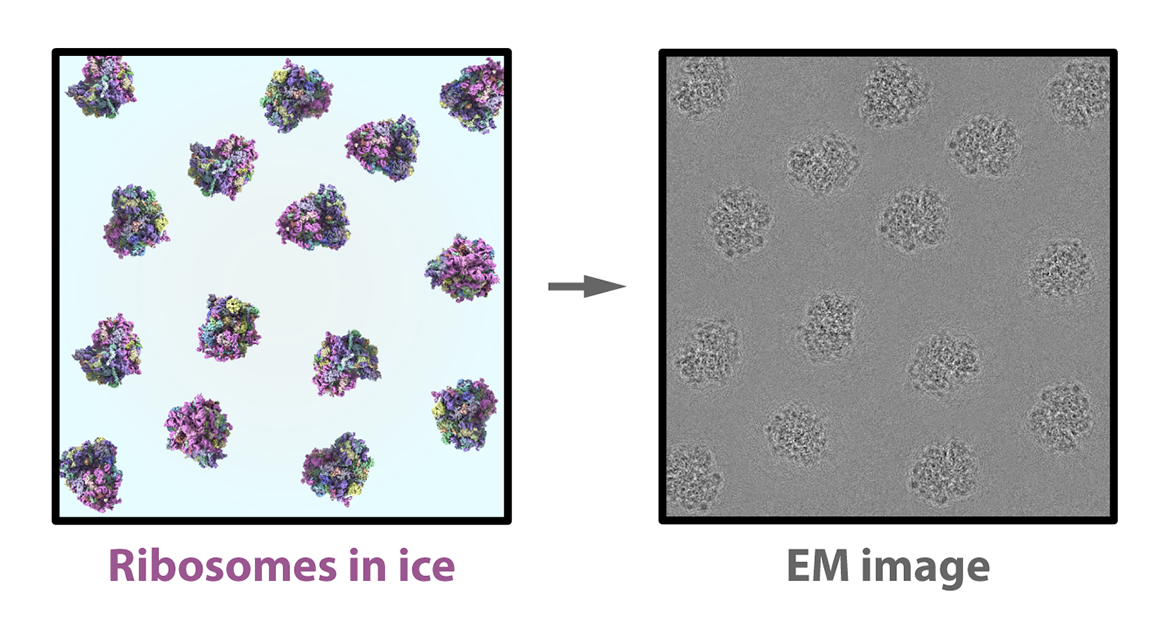
At the computer


What is signal-to-noise?
The thing researchers want to capture with any microscopy technique is the signal: in our case, it’s the structure of the ribosome or protein or other molecule. But unwanted background noise also appears in the image, making it appear grainy — like a photograph taken in the dark of night. The higher the signal-to-noise ratio, the clearer the picture of the structure we’re trying to see.
The challenge posed by taking pictures with cryo-EM is that it is always night, metaphorically speaking, so its images are among the “noisiest” of any imaging technique. That’s largely because a high dosage of electrons, which could boost the signal, would also do too much structural damage to the particles in the sample. The low-dosage electron beam used in cryo-EM results in low-signal, noisy images. To deal with this problem, a cryo-EM expert can mimic what a photographer might do: extend their exposure time. But instead of extending exposure for one particle, the microscopist takes a huge quantity of images of different particles. We can assume these particles take the same shape in the “wild,” and for every orientation the particles appear in, we can start to build a representative image — known as a 2D class average — with an improved signal-to-noise ratio.


Rendering of the Discovery Building, on the UW–Madison campus, depicting the graininess of low signal-to-noise night photography versus the clarity of a higher signal-to-noise image.
An evolving field
To fully realize the potential of cryo-EM, scientists are overcoming its bottlenecks and pushing its boundaries. All of this work goes back to cryo-EM’s founding principle: better visualizing life’s machinery at work in the most natural cellular habitat possible. Scientists are going after even higher resolutions for even smaller molecules and more complex kinds of samples. And above all, they are seeking to better visualize molecular machinery in motion, not just at rest.
These goals require innovation at every step, from sample preparation to image processing. So, for example, scientists are working to develop new ways to freeze molecules before they go under the microscope and new computational methodology for capturing molecules as they move. They are even aiming to view proteins as they do their work inside the cell — known as “in situ structural biology.”
In this effort, a form of cryo-EM called cryogenic electron tomography (cryo-ET for short) is especially exciting. Cryo-ET is the only structural biology method available today that can image everything from molecules to cells to tissues to small organisms in their near-native context. Whereas single-particle analysis takes many frames of many particles on the grid, cryo-ET takes many frames of the same sample as it is tilted under the microscope to capture different parts of its structure. Computational steps align and merge these images, forming a tomogram, or a 3D picture that can illuminate how molecules carry out their work inside the cell.
For example, cryo-ET has revealed how particles of respiratory syncytial virus assemble at the plasma membrane and morph into filaments, creating the most infective form of the virus. RSV (as the virus is widely known) can cause severe respiratory problems, especially in infants and older adults, and understanding how its structure and its power to infect us are related is crucial to developing antiviral treatments. The video below captures how viral RSV components first accumulate inside an infected cell to initiate assembly of a new viral particle, which elongates into the filament shape before narrowing and undergoing “scission,” a process that severs the viral particle from the cell membrane so that it can invade a new host cell.

Image (scale: 1,000 nanometers) and video (scale: 500 nanometers, zeroing in on the area in the white dashed box) where RSV particles assemble and form infective filaments, adapted from a 2018 paper published in the journal Viruses.3
Cryo-ET is also helping structural biologists bridge a gap with light microscopy, in particular a technique that uses fluorescent labels to highlight and more precisely locate features of interest in the crowded, bustling environment of the cell.
Indeed, with increasing access to cutting-edge technology, the future of structural biology lies in combining complementary imaging techniques. For example, scientists are combining single-particle cryo-EM with NMR to leverage their respective advantages with bigger molecule sizes and higher resolution. Others are honing a technique called microcrystal electron diffraction (known as MicroED), which combines cryo-EM with the diffraction approach used in X-ray crystallography to uncover structural details of samples that otherwise resist the crystallization process.
A Brief History of Cryo-EM
So, how did we get here? Let’s take a look back at some key moments in the fascinating history of cryo-EM!
1931: invention of the electron microscope
German scientists Max Knoll and Ernst Ruska (pictured below in 1944) invented the first electron microscope, setting up the possibility of overcoming the limitations imposed by the natural properties of light on what could be seen by modern microscopes. Ruska later won the Nobel Prize in Physics for this work.
1970: three dimensions
Thanks to advances in computation, scientists achieved 3D reconstructions of the human wart and tomato bushy stunt viruses — spherical viruses whose non-helical shape required collecting images from several different views of the particles.4
1975: averaging
In another computational advance, scientists introduced the concept of averaging many 2D images of the same view of the particle before further image processing, a crucial step to reduce the effect of the graininess that marks electron microscopy images of biological samples.5
1984: plunge freezing
Scientists took the first cryo-EM pictures of virus molecules that had been plunged into a cryogenic liquid to trap their particles in a layer of glass-like vitreous ice. This sample preparation step remains largely unchanged to this day.6
2012 and beyond: direct electron detection
With commercial availability of direct electron detection technology, scientists bypassed the need to convert images of molecules from the microscope into lower-resolution photographic film. The resolution revolution was born!
In the years since, the number of high-resolution cryo-EM contributions to the Protein Data Bank, an international repository of molecular structures, has skyrocketed.
In this figure drawn from the paper corresponding to PDB entry 8J5A, for example, the core region of the mouse apoferritin protein is resolved to the level of individual atoms! The red arrow is pointing to a single water molecule.7
2017: Nobel victory
The Nobel Prize for Chemistry was awarded to Jacques Dubochet, Joachim Frank, and Richard Henderson for pioneering contributions to cryo-EM.
Cryo-EM at Wisconsin
Morgridge Institute for Research

In a pivot away from traditional cryo-EM sample preparation, research groups led by Joshua Coon and Tim Grant are using techniques from mass spectrometry to land “molecular elephants” — better known as proteins — directly onto cryo-EM grids instead of plunging those grids into liquid ethane.
The Morgridge Institute for Research is a private, nonprofit biomedical research center located on the UW–Madison campus. Here, the lab of virology Investigator Tim Grant develops and hones cryo-EM methodologies, with a particular focus on overcoming sample preparation bottlenecks and pushing the boundaries of the resolution at which dynamic structures can be solved. Grant is also the co-developer of cisTEM, an open-source software that allows users to take their original cryo-EM images through all the steps necessary to produce 3D reconstructions.
In collaboration with University of Wisconsin–Madison Professor and Morgridge Investigator Josh Coon, the Grant Lab combines cryo-EM with mass spectrometry, an analytical technique based on charging particles and measuring molecular mass. The marriage of these two methods makes it possible to land samples directly onto a matrix and then carefully coat them with an extremely thin layer of amorphous ice, bypassing the plunge-freezing approach that hasn’t significantly changed in the last four decades.8 Using mass spectrometry in the process also makes it possible to separate proteins in heterogeneous mixtures, a complication that has traditionally challenged cryo-EM.
“The next few years are going to be a truly exciting time for the field, and I cannot wait to see what we can all achieve, and where the limits will be.”

Tim Grant
Morgridge Institute for Research
In other collaborations, the Grant Lab illustrates the role cryo-EM plays in major scientific discoveries with implications for human health. With virologist Paul Ahlquist’s lab at Morgridge, they revealed the intricate structure of the replication machinery of a nodavirus model at near-atomic resolution.9 The goal is to identify and understand the structure and function of intracellular replication machinery induced by positive-strand RNA viruses, a massive class of deadly pathogens including the viruses that cause COVID-19, HIV/AIDS, Zika, and Hepatitis C. And as captured in the images above, an ongoing collaboration with Biomolecular Chemistry Professor James Keck’s lab at UW–Madison uses cryo-EM to connect the structure of E. coli bacteria to functions like genome replication.
The lower of two stacked 12-mer rings that make up the crown-shaped replication machinery of a nodavirus. This “proto-crown” (whose distinct functional domains are colorized in the video above) recruits the remaining components needed to synthesize new viral RNA genomes — a step that may be targeted to kill the virus. This project is led by Morgridge Investigator Paul Ahlquist in the John W. And Jeanne M. Rowe Center for Research in Virology. Video: Hong Zhan
University of Wisconsin–Madison
Scientists at UW–Madison are harnessing single-particle cryo-EM and cryo-ET, in combination with other structural biology and microscopy techniques, to discover the structures, mechanisms, and dynamics that underlie health and disease.
UW–Madison Biochemistry Professor and Morgridge Affiliate Investigator Elizabeth Wright, for example, leads a group of scientists who pioneer cryo-imaging technology to explore the architecture of cell systems and host–pathogen interactions. Their group is at the forefront of bringing light and electron microscopy together to study how cells regulate their functions (or malfunction) and how viruses like RSV assemble and replicate.
“The field of cryo-EM continues to evolve. In our labs, we’re pioneering cryo-EM and cryo-ET technologies to enable in situ structural cell biology. With these new tools we are already redefining our mechanistic understanding of neurons, blood cells, and virus-infected cells.”

Elizabeth Wright
University of Wisconsin–Madison

Cryo-EM helps us visualize the molecular machinery fueling the synthesis of telomeres in humans. An accessory protein called CST (colored in purple shades) binds a DNA template (yellow) and positions the central cog in the DNA replication machine (Polα-primase, blue shades) for RNA synthesis. Image: Ci Ji Lim
In Biochemistry Professor Ci Ji Lim’s lab, scientists are also creating an interdisciplinary path to discovery. Their group uses cryo-EM and other single-molecule imaging techniques to study telomeres — the protective “caps” found at the ends our chromosomes. (Imagine the ends of your shoelaces, but made up of repetitive DNA sequences and special proteins.) Telomeres are crucial to the stability of our genome and protecting the ends of our chromosomes from unwarranted damage and repairs.
Scientists are still learning how telomeres are maintained and organized, and how they are made. The Lim Lab found that human telomeres are made using a specialized machinery. The associated structure illuminated by cryo-EM, shown here, is poised to initiate telomere C-strand DNA synthesis, which is crucial not just for maintaining telomeres but also for restarting stalled DNA replication forks across the genome.10
“Cryo-EM has revolutionized how scientists can obtain a three-dimensional model of their biomolecule of interest, especially those that are monstrous in size and jiggle to perform their functions in cells. My lab has benefited from using cryo-EM as a tool to visualize how certain telomeric proteins and enzymes assemble at chromosome ends to complete telomere replication.”

Ci Ji Lim
University of Wisconsin–Madison
And Biochemistry Professor Robert Kirchdoerfer’s group uses cryo-EM to illuminate an important class of proteins called viral polymerases. Coronaviruses such as SARS-CoV, SARS-CoV-2 (the viruses that cause COVID-19), and MERS-CoV use these proteins to make copies of their viral genomes and create instructions to produce viral proteins. Understanding these processes in turn helps us understand how viruses evolve and discover effective antiviral drugs. Cryo-EM plays an important role in revealing the structures underlying these processes. For example, the Kirchdoerfer group used cryo-EM to reconstruct the polymerase of an alphacoronavirus called porcine epidemic diarrhea virus — a global pathogen that is particularly fatal for neonatal piglets. This distant cousin of betacoronaviruses like SARS-CoV-2, the virus that causes COVID-19, is teaching us which parts of the viral polymerase machine are most important for the machine to carry out its function.
“Cryo-EM has the capability to illuminate the chemistry of molecular machines and teach us how those machines work. This technique is an essential part of our cycles of hypothesis testing that complements traditional biology and virology approaches.”

Robert Kirchdoerfer
University of Wisconsin–Madison

The spike proteins characteristic of coronaviruses, including the porcine epidemic diarrhea virus featured in this cryo-EM structure, are critical for the virus’s ability to attach to and enter host cells, and are prime candidates for antiviral therapeutics. Image: Robert Kirchdoerfer & Alysa Zhou
But the power of cryo technology now reaches far beyond any one lab and their collaborators through cryo-EM core facilities. These facilities have revolutionized the availability of cryo-EM technology and are training a new generation of structural biologists. The UW–Madison Cryo-Electron Microscopy Research Center (CEMRC) is a prime example, bringing the power of cryo-EM to the scientific community at UW–Madison and beyond. The center, led by Biochemistry Professor Elizabeth Wright, is home to four cryo-electron microscopes operated by an expert group of scientists who provide technical assistance, training, and consultation on all steps of the cryo-EM process, from sample preparation and data collection to data processing and analysis.
The CEMRC continues UW–Madison’s long history of collaborative discovery and contributions to structural and cell biology, virology, and medicine. The university is currently hosting over 140 research projects at the CEMRC and another related center housed on campus also led by Professor Wright, the Midwest Center for Cryo-Electron Tomography. Scientists use the centers in diverse and innovative ways — for developing drugs and vaccines, for studying human cell structures, and for advancing instrumentation and computational tools. This kind of work has already yielded important achievements, such as enhanced knowledge of SARS-CoV-2, the virus that causes COVID-19.
Works Cited
- Watson, J. D. & Crick, F. H. C. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature 171, 737–738 (1953).
- Duckworth, A. T. et al. Replication fork binding triggers structural changes in the PriA helicase that govern DNA replication restart in E. coli. Nat Commun 14, 2725 (2023).
- Ke, Z. et al. The Morphology and Assembly of Respiratory Syncytial Virus Revealed by Cryo-Electron Tomography. Viruses 10, 446 (2018).
- Crowther, R. A., Amos, L. A., Finch, J. T., De Rosier, D. J. & Klug, A. Three Dimensional Reconstructions of Spherical Viruses by Fourier Synthesis from Electron Micrographs. Nature 226, 421–425 (1970).
- Frank, J. Averaging of low exposure electron micrographs of non-periodic objects. Ultramicroscopy 1, 159–162 (1975).
- Adrian, M., Dubochet, J., Lepault, J. & McDowall, A. W. Cryo-electron microscopy of viruses. Nature 308, 32–36 (1984).
- Maki-Yonekura, S., Kawakami, K., Takaba, K., Hamaguchi, T. & Yonekura, K. Measurement of charges and chemical bonding in a cryo-EM structure. Commun Chem 6, 1–8 (2023).
- Westphall, M. S. et al. Cryogenic Soft Landing Improves Structural Preservation of Protein Complexes. Anal. Chem. 95, 15094–15101 (2023).
- Zhan, H. et al. Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching. Proceedings of the National Academy of Sciences 120, e2217412120 (2023).
- He, Q. et al. Structures of the human CST-Polα–primase complex bound to telomere templates. Nature 608, 826–832 (2022).







