Simple beginnings: the plentiful planarian
When Phil Newmark landed in Barcelona 19 years ago—carrying a cooler full of razor blades, latex gloves and thermoses—he first looked for a butcher shop.
He bought sheep’s liver, chopped it into little pieces, and roamed the city hunting through the public fountains.
His quarry? Planarians. Small, nondescript flatworms that live in fresh water and have amazing regenerative properties.
Newmark and his research colleague needed to replenish their stock of the organisms for their lab and knew populations of planarians lived in one of Barcelona’s fountains.
“Usually you’d see tons of them in the fountain. We put on our latex gloves—because you really didn’t want to touch the water or its contents—and picked up stones and leaves looking for planarians,” Newmark says. “We were there for hours. Between the two of us we only found one planarian.”
They used fishhooks and twine to leave behind pieces of liver, a tantalizing treat, and came back the next day to find thousands of the worms.

Today, Newmark can count even more planarians in his laboratory at the Morgridge Institute for Research. As a principal investigator in regenerative biology, his lab studies these humble planarians because of their remarkable ability to regenerate virtually any part of their body from scratch.
This magnificent regeneration comes a from a population of pluripotent stem cells called neoblasts, says Melanie Issigonis, an assistant scientist in the Newmark Lab.
As long as an amputated segment contains a single stem cell, the entire organism will regenerate. Simply put, split a planarian into 10 separate pieces and you’d end up with 10 worms.
“Planarians are masters of regeneration, with virtually unlimited capacity to regenerate tissues lost to injury or aging,” says Issigonis. “This ability to regenerate comes from a population of highly potent stem cells.”
What’s especially interesting is the planarian’s ability to regenerate extends to the germline. Germ cells are considered to be immortal because they’re responsible for making reproductive cells, sperm and eggs. They create the cells that produce new life, and can be passed on to the next generation.
Planarians form their germ cells in the same way we do, and the Newmark Lab is studying this process to better understand infertility in humans.
“It’s an interesting question from a basic biology standpoint, but there are also important clinical implications,” Issigonis says. “Infertility is a large and growing problem in our society. Around 15 percent of couples have fertility issues, and that number keeps getting worse.”
Planarians are masters of regeneration, with virtually unlimited capacity to regenerate tissues lost to injury or aging. This ability to regenerate comes from a population of highly potent stem cells.

Melanie Issigonis
Assistant Scientist, Regenerative Biology
The planarian’s parasitic cousin
While planarians are carnivores—remember Newmark attracted his worms with sheep’s liver—in the wild they stick to small prey like aquatic insects and snails. Schistosomes, on the other hand, have more sinister tastes.
Schistosomes, flatworm cousins to planarians, are human parasites. They cause schistosomiasis, a neglected tropical disease affecting hundreds of millions of people in Africa, Asia and parts of South America. While mortality rates vary widely, the World Health Organization estimates that as many as 200,000 people die annually from the disease.
The Newmark lab has been a leader in identifying similar traits shared between planarians and their much more sinister cousins. Schistosomes and planarians both contain banks of stem cells, and share many genes that are only found in the flatworms.
“Applying what we’ve learned and developed for the planarian system to the parasite helps us understand these creatures that are really difficult to study and directly impact human health,” Newmark says.
As a parasite, schistosomes depend on their hosts for survival. They asexually reproduce in an intermediate host – a species of snail – before infecting humans or other mammals.
An infected snail releases thousands of cercariae, the swimming, tadpole-like stage of the parasite that is infectious to humans. The cercariae can only survive for 48 hours without a host, so their only goal is to find someone to infect.
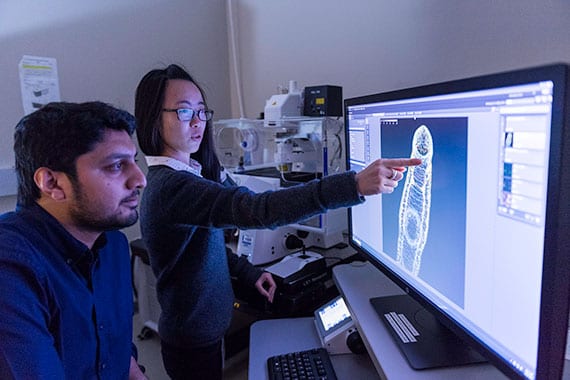 Jiarong Gao points at the schistosome brain in an adult worm.
Jiarong Gao points at the schistosome brain in an adult worm.  An infected snail releases the swimming, tadpole-like stage of the parasite that is infectious to humans.
An infected snail releases the swimming, tadpole-like stage of the parasite that is infectious to humans. 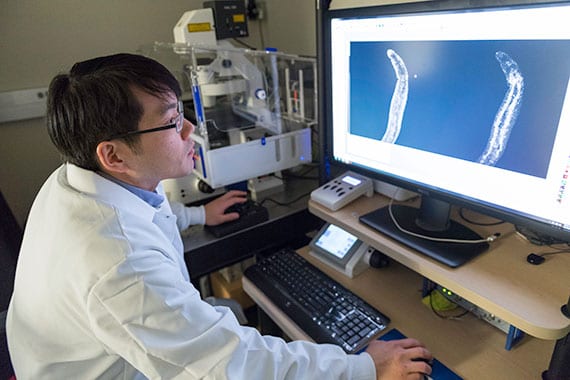 Jayhun Lee, a postdoctoral fellow in the Newmark Lab, looks at the gut of a juvenile schistosome.
Jayhun Lee, a postdoctoral fellow in the Newmark Lab, looks at the gut of a juvenile schistosome. Jiarong Gao, a graduate student researcher in the Newmark Lab focusing on the schistosomes work, says cercariae find humans and other mammals by sensing their body heat and chemicals in the skin. The parasites burrow into the skin and travel through the blood vessels until they end up near the liver, where they make themselves at home.
“They feed on blood cells, become sexually mature and pair up,” Gao says. “Each pair can release hundreds of eggs every day, which either get passed out of the body through feces and urine or get stuck in the human’s liver.”
The worms can live for decades, maybe longer, continuing to lay eggs and wreak havoc on the liver until the host eventually dies.
Currently, there’s only a single drug to combat schistosomiasis, and it has significant limitations and no way to prevent against reinfection. The Newmark Lab is hoping that by better understanding the basic biology of the parasite, new opportunities for treatment will emerge.
The immortality of tapeworms
Tapeworms, a more notorious parasite, are also known for feats of regeneration, relying largely on a bank of stem cells to fuel their growth.
“I think we should study the monsters in our midst,” says Tania Rozario, a postdoctoral researcher in the Newmark Lab. “They are all around us and they have remarkable biology and physiology that evolution has already selected for. It just behooves us to figure out how they work.”
Humans or animals can be infected by ingesting a tapeworm or its larva via undercooked meat or from food prepared in unsanitary conditions. The head of a tapeworm, its scolex, attaches itself to the intestine of its host, where it stays indefinitely.
The body is composed of unique segments, each of which have independent reproductive systems. The neck region of the head is continually producing new segments—as long as the scolex is in place, the worm will continue to grow. The older end segments of the tail, essentially bags of fertilized eggs, will drop off and leave the body through feces.
I think we should study the monsters in our midst. They are all around us and they have remarkable biology and physiology that evolution has already selected for. It just behooves us to figure out how they work.

Tania Rozario
Postdoctoral Fellow, Regenerative Biology
The largest species of tapeworm, the whale tapeworm, can grow to more than 100 feet long. But it’s difficult to know the true limits of size, since the tapeworm is continually growing and shedding segments. No matter how long a tapeworm gets, its head—where the regenerative power lies—remains the same small size.
The tapeworm’s growth is remarkable, rivaling any we have seen in life, Rozario says, including cancerous growth or embryonic development. It seems like the worms themselves don’t age.
“They might be immortal,” Rozario says. “If you can artificially transplant them so that they’re not restricted by the life span of the host, they can live for many, many years.”
Rozario says that in studying this organism there will undoubtedly be discoveries not translatable beyond tapeworms. But findings that reveal biological principles could be extrapolated to other organisms—lessons that are applicable across all stem cell based systems.
“Models for biology often have an unknown scope, but I think we should embrace that,” Rozario says. “If we could see the end of the road, we would be there.”
Mexican ‘water monster’ is regeneration king
The axolotl, a Mexican salamander, is an amphibious master of regeneration.
The creature, sometimes referred to as a “water monster,” has wide, lidless eyes, short legs and a head framed by feathery external gills. Its front limbs each sport four finger-like digits, while the back limbs each have five.
And should the axolotl ever lose a digit—or even a leg or its tail—it can grow it right back.
“Among vertebrate tetrapods—things with four limbs—the axolotl is one of the best regenerators out there,” says Jeffrey Nelson, a former postdoctoral fellow in regenerative biology at the Morgridge Institute. “It can regenerate its limbs, tail, parts of the eye and heart, even its central nervous system.”
Nelson studies these creatures to better understand the regenerative ability of vertebrates, a more complex group of animals than simple invertebrates like worms.
It may be hard to imagine growing back your lost finger or leg, but for these water monsters, it’s an ability they excel at.
The axolotl can go through its entire life cycle and reproduce while being in a juvenile aquatic state. This means that instead of ditching the gills and transitioning into a semi-aquatic land animal like other salamanders, it stays aquatic—the state in which its regeneration potential is most powerful.
Among vertebrate tetrapods—things with four limbs—the axolotl is one of the best regenerators out there. It can regenerate its limbs, tail, parts of the eye and heart, even its central nervous system.

Jeffrey Nelson
Postdoctoral Fellow, Regenerative Biology
For researchers at the Morgridge Institute, the hunt is on to identify the specific genes playing a role in the axolotl’s limb regeneration. In defining that growth process, they can also explore if this quirky salamander provides a regeneration recipe of sorts that can be translated to other species.
One challenge is understanding the immensity of the axolotl’s genome. For a relatively simple aquatic creature, it’s genome is more than 10 times larger than a human’s. The secret of regeneration, researchers believe, may lie somewhere in the labyrinth of genes we don’t possess.
“Limb regeneration is a pretty remarkable process, and we know bits and pieces of how it is functioning,” Nelson says. “We don’t know overall how it’s all connected and orchestrated to come together. It’s something important to understand if we’re ever going to become better regenerators as mammals.”

Another alluring mystery is the axolotl’s ability to stop uncontrolled cell growth. Seemingly like a light switch, these organisms can trigger regeneration and stop it, preventing uncontrolled or cancerous growth.
For example, within the first day or so of an injury, the axolotl displays an increased presence of cancer-related oncogenes. They’re the red flag, signaling that an injury has occurred and the regeneration process begins.
Surprisingly, Nelson says, even in the presence of these cancer-related genes, the axolotl doesn’t get cancer at very high rates at all.
“If you were trying to regrow a tissue or limb, you’d want to very tightly control that process so you don’t make an organism develop cancer instead,” Nelson adds.
Somehow the axolotl manages to find this balance between successful regeneration and going too far, the formation of cancer.
While we may never have the ability to regrow our own fingers and toes, the axolotl could still unravel mysteries around simpler wound healing and identify genes important to stem cell based organisms—salamanders and humans alike.
Yeast enables exploration of 'powerhouse of the cell'
Yeast may seem more at home in a baker’s kitchen or a brewery, but this simple, single-celled organism has vast human health applications.
In particular, these tiny organisms can tell us a lot about mitochondria, the “powerhouse of the cell” responsible for energy production. More than 150 disorders and diseases have been linked to mitochondrial dysfunction, including Parkinson’s, diabetes and Alzheimer’s.
Danielle Lohman, a graduate research assistant in Dave Pagliarini’s lab, part of the Morgridge Metabolism Theme, recalls a case study from the early 2000s of a young boy with coenzyme Q10 deficiency leading to multi-systemic disease, conditions that affect many systems in the body. His rare, incurable case was linked to a dysfunction in a specific gene called coenzyme Q9, or COQ9.
At the time, doctors were unable to develop a successful therapy or treatment, but research today in the field of metabolism is working to unlock these mysteries.
“My job is to figure out the purpose of that gene,” Lohman says. “One of the ways researchers do this is to study the same mutation that was in the patient, but in yeast.”

Why yeast? Humans and yeast actually share thousands of genes, of which several hundred—discovered so far—have consistent functions across the two systems. Zeroing in on specific gene functions or mutations in yeast can be applicable to humans as well.
Think of a cell as a machine, like a car engine, says Lohman. You know when your car isn’t working and you may know which part is responsible. But unless you know how the entire engine works and what that part is doing, you can’t fix the problem.
In humans, it’s quite difficult to lift up the hood and take a peek at the engine. That’s where yeast comes in.
Studies in yeast largely resolve the mystery of the genetic architecture by revealing functions of specific traits. Lohman says that yeast cells are not only easier to study than human cells, they’re easier to manipulate genetically. With high-tech laboratory equipment, scientists can remove genes like COQ9, or make specific mutations to study and understand what happens when this protein is altered.
“Model organisms accelerate the discovery process,” says Kyle Robinson, a graduate research assistant in metabolism at the Morgridge Institute. “Yeast are stripped down to the basics. If you want to understand a complicated biological process, it helps to study it at a basic level first.”
Like humans, yeast belongs to a group called eukaryotes—organisms consisting of cells with a distinct nucleus containing genetic material in the form of chromosomes. Yeast is a particularly simple organism – and also the first to have its entire genome sequenced – making it an excellent screening tool for health applications.
The conciseness of the yeast genome makes it much easier to study. In humans, with our much larger genome, there may be four different gene products responsible for one task that is carried out by a single gene product in yeast.
Model organisms accelerate the discovery process. Yeast are stripped down to the basics. If you want to understand a complicated biological process, it helps to study it at a basic level first.

Kyle Robinson
Graduate Research Assistant, Metabolism
Scientists can grow billions of yeast cells overnight. This quick, easy growth and simple genome makes this model organism ideal for genetic screens, which are used to identify changes in genes. They’ve helped uncover how cells go through the cell cycle, DNA replication, and even the process of cell division.
Scientists must make sense of fundamental processes—like gene regulation—in a model organism such as yeast, before knowing enough to fully decipher the process in humans.
The battle against microbial overlords
In the fight against “microbial overlords”—the viruses that invade our daily lives—being able to see and understand the battleground is a huge advantage.
“It’s like this epic battle between us and our microbial overlords on a day-to-day basis,” says Desirée Benefield, a postdoctoral researcher in virology at the Morgridge Institute.
Benefield works in the lab of Paul Ahlquist, an HHMI and Morgridge investigator, where she investigates viral replication in host cells using high-tech microscopes.
The Ahlquist team is seeking to understand genome replication for positive strand RNA viruses, the largest genetic class of viruses that includes many serious human pathogens such as the Zika, Dengue, SARS, and Chikungunya viruses. Their aim is to expand our knowledge of these viruses in the hopes to improve human health.
To do so, the team uses several models including Flock house virus (FHV), an insect virus unable to transmit to humans. FHV’s incredibly small genome makes it an ideal model for research, especially in understanding the key events in a virus’s life cycle with a specific focus on genome replication.
If we can slow the process down by targeting the proteins associated with viral replication, that might buy the immune system a little time to really mount up its defense while the virus is inhibited.

Desirée Benefield
Postdoctoral Researcher, Virology
While the human immune system is pretty robust in most cases, viruses replicate so rapidly that they can easily overcome our immune defenses. If we can understand the intricacies of these events, we may be able to slow, stop or severely limit the impact of pathogens.
“If we can slow the process down by targeting the proteins associated with viral replication,” Benefield says, “that might buy the immune system a little time to really mount up its defense while the virus is inhibited.”
Benefield uses techniques like cryoelectron microscopy—in which a sample is embedded in vitreous ice, frozen in a near native state—to study cellular organelles that have been co-opted by the virus for its replication. The sample, in this case, is mitochondria from Drosophila, fruit fly, cells.
FHV viral genome replication requires the formation of replication compartments, little spherules, in the mitochondria of an infected host cell. Benefield wants to understand the overall architecture of these complexes, how they are organized, and how they facilitate replication.
The more we know about how viruses are replicating their genome, the better scientists can identify targets for drug therapy and develop antivirals in the future.
Microscopy is one of the tools that will help us get there.
“I love being on a microscope, I love all of the processes involved,” Benefield says. “When it all comes together it’s one of the best feelings in the world. A eureka moment, even if it’s a small eureka moment, it’s wonderful.”
Zebrafish benefits are transparent
Model organisms each have their own characteristics or peculiarities that make them ideal for studying biology. When it comes to zebrafish, you can see right through them.
Zebrafish embryos are transparent, giving researchers an inside look at the development of internal structures. These windows help expand our knowledge of these vertebrate systems and open up the possibility for advances in human health.
“Zebrafish start off as little one-millimeter sized balls, sort of like caviar,” says Jan Huisken, director of medical engineering at Morgridge. “They grow very well in a petri dish, which makes them easy to image, and then grow to look like a regular fish—with black stripes.”
It was the zebrafish’s developing cardiovascular system that caught his eye. In particular, the heart is a fascinating organ to image – it’s located close to the surface of the developing zebrafish embryo where it can be easily imaged and manipulated.
“We can optically pace or change the frequency of the heart,” Huisken says. “We can study a lot of developmental, but also functional processes in the cardiovascular system.”
But, this is a race against the clock. The fish’s tiny heart starts beating at around 24 hours after fertilization. From there, a small window of time – about three days – gives researchers a chance to image the formation of major organs in the translucent embryo. Pigment also starts to appear—the stripes that put the ‘zebra’ in zebrafish—losing some of the transparency that makes the fish so appealing.

Huisken’s development of light sheet microscopy, a novel fluorescent imaging technique, has led to major breakthroughs in imaging early development of several major model organisms, including the zebrafish. The technique gently images fast biological processes that are extremely sensitive, in vivo.
His microscope uses multi-view reconstruction to look at an embryo from different sides and then stitch all the data together. The technology can gather snapshots in time and image a developing fish, Huisken says.
Many microscopes in the Huisken lab are optimized for the fish, taking special care that the specimen actually survives the experiment. Almost all of the tools used are developed and tailored around the zebrafish. It is this flexibility and customizability that makes light sheet microscopy so powerful.
“We feel very proud that everything we do is something where the fish can be taken out of the microscope at the end of the experiment and live on its life,” Huisken says. “We have the optimal setup to image a living organism.”
The range of Huisken’s lab is nicely illustrated by fish on one end swimming in a tank, while on the other end a computer model shows cardiac output in a 24-hour-old zebrafish.
“We have our own fish, and we breed them, prepare them, select them for imaging. We build and program the microscope, take and analyze the data, and use that information to better understand embryo development,” Huisken says.
With state-of-the-art imaging tools and ample supply of zebrafish embryos for the lab to study, Huisken hopes to eventually take biomedical imaging to an even higher level—where we can image living mammalian tissue for an extended period of time.
The next model organism: Ourselves
Model organisms have an important place in understanding biology, but there will always be limitations. These creatures can only tell us so much about human biology.
About 90 percent of new drug candidates fail during human clinical trials because they are ineffective (the majority) or unsafe, according to the NIH. Potential drugs only reach the human trial stage if they’ve passed certain hurdles in bench or animal testing, so there must still be some disconnect between animal models and humans.
Around 100,000, mostly untested, chemical compounds are used in commerce today with new ones added every year. Current toxicity screening tests use multi-generational rat studies and cost about $1 million to test one chemical.
In reality, some questions in biology can only be answered by ourselves.
Our Brains: Tissues on a chip
A team led by James Thomson, director of regenerative biology at Morgridge, is working to bring humans—or at least our cells—into the forefront of biomedical science.
His lab is part of a National Institutes of Health (NIH) multi-site project to create models of human tissue in a dish. These “tissue chips” or organoids require human stem cells to develop 3-D cultures that mimic the structure and function of human organs like the heart, brain and liver.
One Thomson Lab research project focused on the brain. They produced a model of human neural tissue with features that resemble the developing human brain. Each chip is about one-fifth the circumference of a dime, yet is sophisticated enough to imitate the functions of human tissue in the body.
Using machine learning and robotics technology, the team developed a system for high-throughput screening of chemicals to identify neurotoxins. This means they could screen lots of chemicals or drugs at once to discover which might be harmful to the developing brain.
With tissue chips, drugs could be tested on actual human cells and provide greater predictivity to which are most likely to be successful in clinical trials and go on to patient use.
Tackling pancreatic cancer: personalized medicine
For Melissa Skala, cancer can be stopped.
We want to be able to predict the best patient therapy before the patient actually receives any drugs. We can determine the best treatment strategy for each patient by essentially measuring the metabolism of the cells in response to the drugs that they may get.

Melissa Skala
Principal Investigator, Medical Engineering
Skala, a principal investigator in medical engineering at Morgridge, is working with human cells to develop novel clinical treatments against cancer. Currently, cancer and its potential treatments can be highly unpredictable — what fails in one patient may succeed in another.
“I think scientists in general like to solve problems,” Skala says. “They like to put together puzzles. We are motivated by new technologies because we’re engineers, but we’re also motivated by impactful work.”
Skala is working to develop a range of strategies to help doctors and patients determine the most effective treatments against cancer. Her eyes are tightly focused on pancreatic cancer, a disease with the highest mortality rate of any cancer. Patients have few options and because of its low survival rate, it’s a cancer that remains drastically under-researched.
Skala uses samples of a patient’s cancerous tumor to create a set of micro-tumors, with the cells kept alive in collagen. These micro-tumors are given a variety of treatments—the same options available to patients—to help determine which treatment may be most effective in a specific case.
The potential impact and application of this work is a great example of “personalized medicine,” the buzzwords representing the latest aspiration for healthcare. It’s becoming more apparent, especially in cancer, that there’s no one-size-fits-all treatment.
“We want to be able to predict the best patient therapy before the patient actually receives any drugs,” says Skala. “We want to minimize toxicities from treatments that don’t work, and we want to use treatments that do work. We can determine the best treatment strategy for each patient by essentially measuring the metabolism of the cells in response to the drugs that they may get.”
New advances are helping shift the scientific process, bringing our cells into a new paradigm for discovery. While we may never replace all model organisms, these creatures – in addition to our own selves – are helping scientists better understanding the complexity of biology to drive tomorrow’s cures.

